Api regional
Summary
All regional effect methods have a similar interface and workflow:
- create an instance of the regional effect method you want to use
- (optional)
.fit()to customize the method .summary()to print the partition tree found for each feature.plot()to plot the regional effect of a feature at a specific node.eval()to evaluate the regional effect of a feature at a specific node at a specific grid of points
Usage
# set up the input
X = ... # input data
predict = ... # model to be explained
jacobian = ... # jacobian of the model
-
Create an instance of the regional effect method you want to use:
effector.RegionalPDP(data=X, model=predict)effector.RegionalRHALE(data=X, model=predict, model_jac=jacobian)effector.RegionalShapDP(data=X, model=predict)effector.RegionalALE(data=X, model=predict)effector.DerPDP(data=X, model=predict, model_jac=jacobian) -
Customize the regional effect method (optional):
.fit(features, **method_specific_args)This is the place for customization
The
.fit()step can be omitted if you are ok with the default settings; you can directly call the.summary(),.plot(), or.eval()methods. However, if you want more control over the fitting process, you can pass additional arguments to the.fit()method. Check the Usage section below and the method-specific documentation for more information.Usage
# customize the space partitioning algorithm space_partitioner = effector.space_partitioning.Best( min_heterogeneity_decrease_pcg=0.3, # percentage drop threshold (default: 0.1), max_split_levels=1 # maximum number of split levels (default: 2) ) r_method.fit( features=[0, 1], # list of features to be analyzed space_partitioner=space_partitioner # space partitioning algorithm (default: effector.space_partitioning.Best) ) -
Print the partition tree found for each feature in
features:.summary(features)Usage
features = [...] # list of features to be analyzed r_method.summary(features)Example Output
Feature 3 - Full partition tree: 🌳 Full Tree Structure: ─────────────────────── hr 🔹 [id: 0 | heter: 0.43 | inst: 3476 | w: 1.00] workingday = 0.00 🔹 [id: 1 | heter: 0.36 | inst: 1129 | w: 0.32] temp ≤ 6.50 🔹 [id: 3 | heter: 0.17 | inst: 568 | w: 0.16] temp > 6.50 🔹 [id: 4 | heter: 0.21 | inst: 561 | w: 0.16] workingday ≠ 0.00 🔹 [id: 2 | heter: 0.28 | inst: 2347 | w: 0.68] temp ≤ 6.50 🔹 [id: 5 | heter: 0.19 | inst: 953 | w: 0.27] temp > 6.50 🔹 [id: 6 | heter: 0.20 | inst: 1394 | w: 0.40] -------------------------------------------------- Feature 3 - Statistics per tree level: 🌳 Tree Summary: ───────────────── Level 0🔹heter: 0.43 Level 1🔹heter: 0.31 | 🔻0.12 (28.15%) Level 2🔹heter: 0.19 | 🔻0.11 (37.10%) -
Plot the regional effect of a feature at a specific node:
.plot(feature, node_idx)Usage
feature = ... node_idx = ... r_method.plot(feature, node_idx, **plot_specific_args)Output
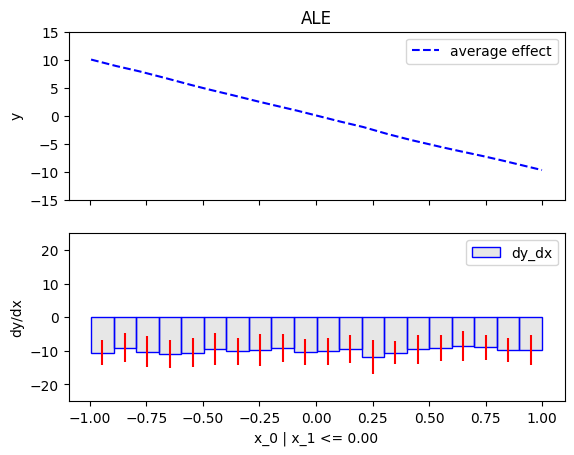
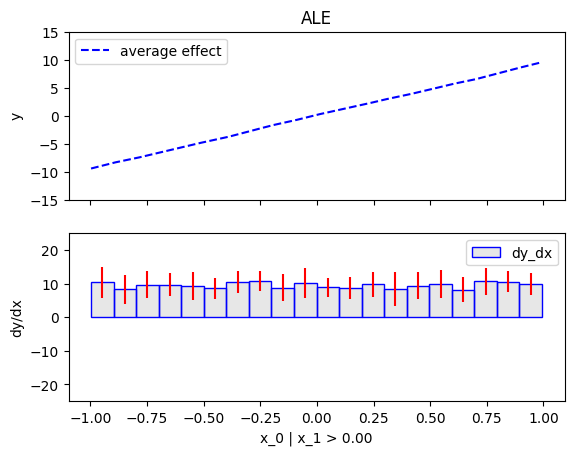
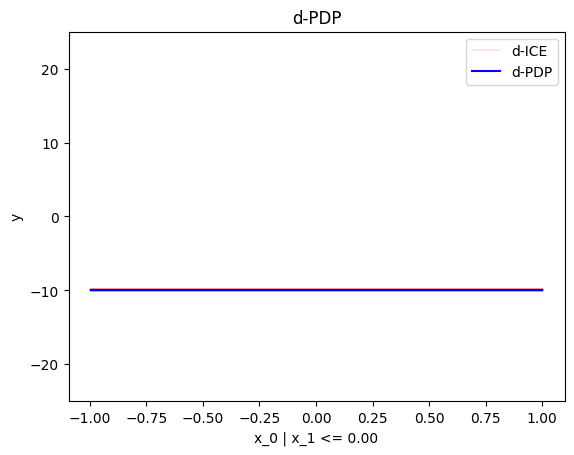
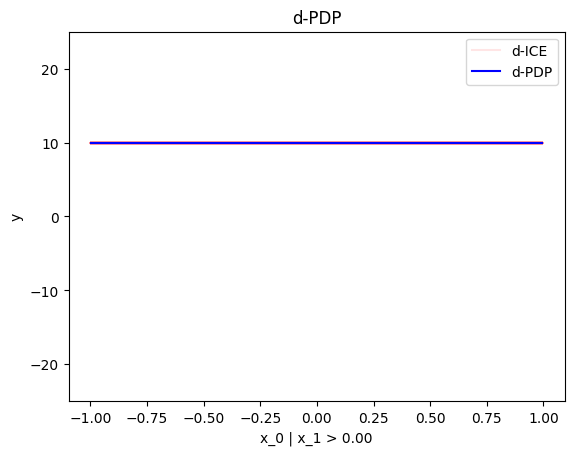
node_idx=1: \(x_1\) when \(x_2 \leq 0\)node_idx=2: \(x_1\) when \(x_2 > 0\)r_method.plot(0, 1)r_method.plot(0, 2)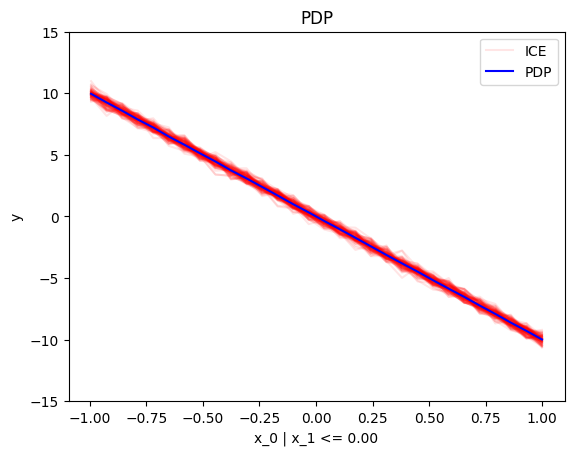
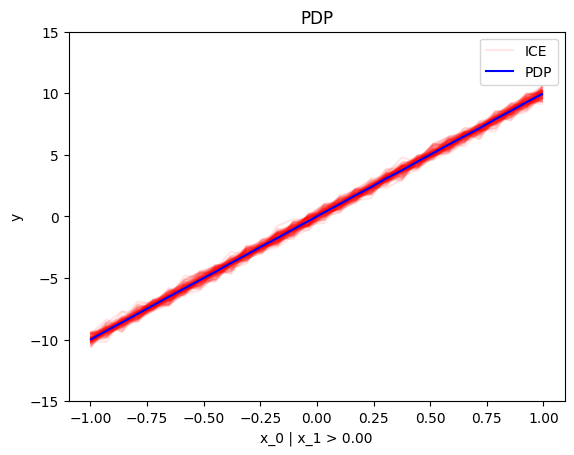
node_idx=1: \(x_1\) when \(x_2 \leq 0\)node_idx=2: \(x_1\) when \(x_2 > 0\)r_method.plot(0, 1)r_method.plot(0, 2)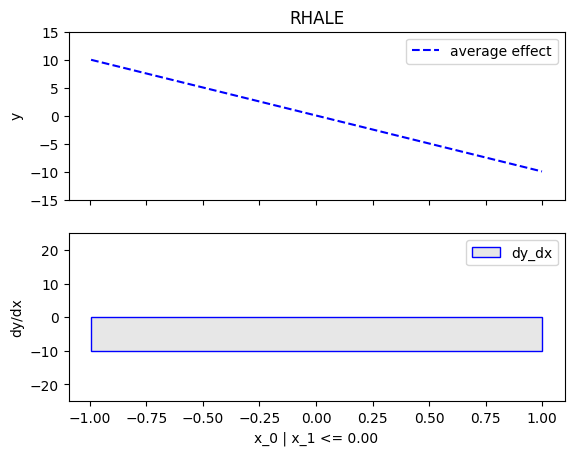
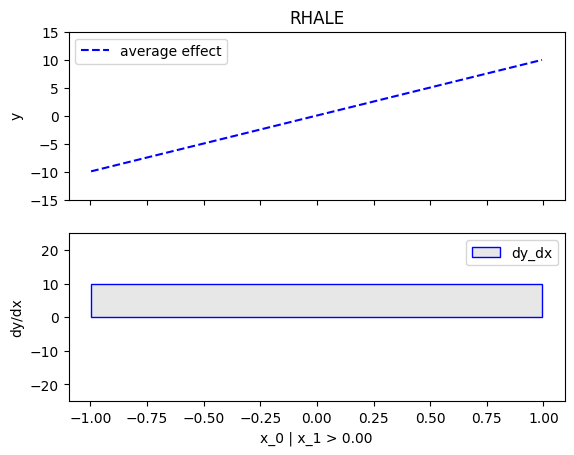
node_idx=1: \(x_1\) when \(x_2 \leq 0\)node_idx=2: \(x_1\) when \(x_2 > 0\)r_method.plot(0, 1)r_method.plot(0, 2)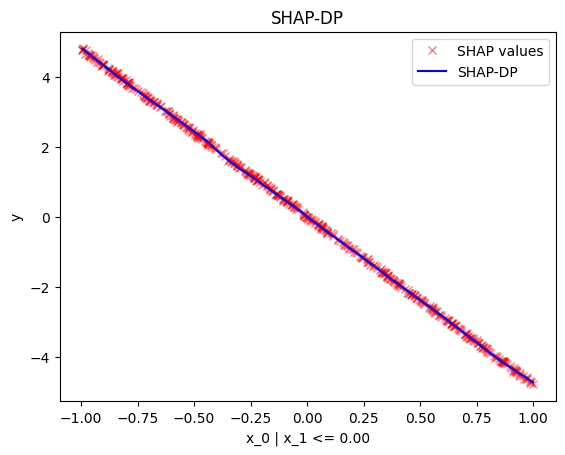
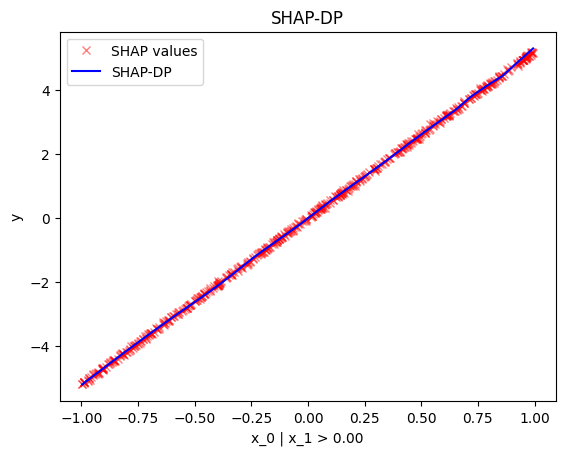
-
Evaluate the regional effect of a feature at a specific node at a specific grid of points:
.eval(feature, node_idx, xs)Usage
# Example input feature = ... # feature to be analyzed node_idx = ... # node index xs = ... # grid of points to evaluate the regional effect, e.g., np.linspace(0, 1, 100)y, het = r_method.eval(feature, node_idx, xs)
API
Constructor for the RegionalEffect class.
Methods:
| Name | Description |
|---|---|
eval |
|
summary |
|
Source code in effector/regional_effect.py
16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 | |
eval(feature, node_idx, xs, heterogeneity=False, centering=True)

This is a common method for all regional effect methods, so use the arguments carefully.
centering=Trueis a good option for most methods, but not for all.DerPDP, usecentering=False[RegionalPDP, RegionalShapDP], it depends on you[RegionalALE, RegionalRHALE], usecentering=True
The heterogeneity argument changes the return value of the function.
- If
heterogeneity=False, the function returnsy - If
heterogeneity=True, the function returns a tuple(y, std)
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
feature
|
int
|
index of the feature |
required |
node_idx
|
int
|
index of the node |
required |
xs
|
ndarray
|
horizontal grid of points to evaluate on |
required |
heterogeneity
|
bool
|
whether to return the heterogeneity.
|
False
|
centering
|
Union[bool, str]
|
whether to center the regional effect. The following options are available:
|
True
|
Returns:
| Type | Description |
|---|---|
Union[ndarray, Tuple[ndarray, ndarray]]
|
the mean effect |
Source code in effector/regional_effect.py
203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 | |
summary(features, scale_x_list=None)

Example output
Feature 3 - Full partition tree:
🌳 Full Tree Structure:
───────────────────────
hr 🔹 [id: 0 | heter: 0.43 | inst: 3476 | w: 1.00]
workingday = 0.00 🔹 [id: 1 | heter: 0.36 | inst: 1129 | w: 0.32]
temp ≤ 6.50 🔹 [id: 3 | heter: 0.17 | inst: 568 | w: 0.16]
temp > 6.50 🔹 [id: 4 | heter: 0.21 | inst: 561 | w: 0.16]
workingday ≠ 0.00 🔹 [id: 2 | heter: 0.28 | inst: 2347 | w: 0.68]
temp ≤ 6.50 🔹 [id: 5 | heter: 0.19 | inst: 953 | w: 0.27]
temp > 6.50 🔹 [id: 6 | heter: 0.20 | inst: 1394 | w: 0.40]
--------------------------------------------------
Feature 3 - Statistics per tree level:
🌳 Tree Summary:
─────────────────
Level 0🔹heter: 0.43
Level 1🔹heter: 0.31 | 🔻0.12 (28.15%)
Level 2🔹heter: 0.19 | 🔻0.11 (37.10%)
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
features
|
List[int]
|
indices of the features to summarize |
required |
scale_x_list
|
Optional[List]
|
list of scaling factors for each feature
|
None
|
Source code in effector/regional_effect.py
279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 | |
effector.regional_effect_ale.RegionalALE(data, model, nof_instances=100000, axis_limits=None, feature_types=None, cat_limit=10, feature_names=None, target_name=None)
Bases: RegionalEffectBase
Initialize the Regional Effect method.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
ndarray
|
the design matrix, |
required |
model
|
callable
|
the black-box model,
|
required |
axis_limits
|
Union[None, ndarray]
|
Feature effect limits along each axis
When possible, specify the axis limits manually
Their shape is |
None
|
nof_instances
|
Union[int, str]
|
Max instances to use
|
100000
|
feature_types
|
Union[list, None]
|
The feature types.
|
None
|
cat_limit
|
Union[int, None]
|
The minimum number of unique values for a feature to be considered categorical
|
10
|
feature_names
|
Union[list, None]
|
The names of the features
|
None
|
target_name
|
Union[str, None]
|
The name of the target variable
|
None
|
Methods:
| Name | Description |
|---|---|
fit |
Find subregions by minimizing the ALE-based heterogeneity. |
plot |
|
Source code in effector/regional_effect_ale.py
232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 | |
fit(features, candidate_conditioning_features='all', space_partitioner='best', binning_method='fixed', points_for_mean_heterogeneity=30)
Find subregions by minimizing the ALE-based heterogeneity.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
features
|
Union[int, str, list]
|
for which features to search for subregions
|
required |
candidate_conditioning_features
|
Union[str, list]
|
list of features to consider as conditioning features |
'all'
|
space_partitioner
|
Union[str, Best]
|
the space partitioner to use |
'best'
|
binning_method
|
Union[str, Fixed]
|
must be the Fixed binning method
|
'fixed'
|
points_for_mean_heterogeneity
|
int
|
number of equidistant points along the feature axis used for computing the mean heterogeneity |
30
|
Source code in effector/regional_effect_ale.py
328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 | |
plot(feature, node_idx, heterogeneity=True, centering=True, scale_x_list=None, scale_y=None, y_limits=None, dy_limits=None)
Source code in effector/regional_effect_ale.py
391 392 393 394 395 396 397 398 399 400 401 402 403 404 | |
effector.regional_effect_ale.RegionalRHALE(data, model, model_jac=None, data_effect=None, nof_instances=100000, axis_limits=None, feature_types=None, cat_limit=10, feature_names=None, target_name=None)
Bases: RegionalEffectBase
Initialize the Regional Effect method.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
ndarray
|
the design matrix, |
required |
model
|
Callable
|
the black-box model,
|
required |
model_jac
|
Optional[Callable]
|
the black-box model's Jacobian,
|
None
|
data_effect
|
Optional[ndarray]
|
The jacobian of the
When possible, provide the Jacobian directly Computing the jacobian on the whole dataset can be memory demanding. If you have the jacobian already computed, provide it directly to the constructor. |
None
|
axis_limits
|
Optional[ndarray]
|
Feature effect limits along each axis
When possible, specify the axis limits manually
Their shape is |
None
|
nof_instances
|
Union[int, str]
|
Max instances to use
|
100000
|
feature_types
|
Optional[List]
|
The feature types.
|
None
|
cat_limit
|
Optional[int]
|
The minimum number of unique values for a feature to be considered categorical
|
10
|
feature_names
|
Optional[List]
|
The names of the features
|
None
|
target_name
|
Optional[str]
|
The name of the target variable
|
None
|
Methods:
| Name | Description |
|---|---|
fit |
Find subregions by minimizing the RHALE-based heterogeneity. |
plot |
|
Source code in effector/regional_effect_ale.py
17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 | |
fit(features='all', candidate_conditioning_features='all', space_partitioner='best', binning_method='greedy', points_for_mean_heterogeneity=30)
Find subregions by minimizing the RHALE-based heterogeneity.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
features
|
Union[int, str, list]
|
for which features to search for subregions
|
'all'
|
candidate_conditioning_features
|
Union[str, list]
|
list of features to consider as conditioning features |
'all'
|
space_partitioner
|
Union[str, Best]
|
the space partitioner to use |
'best'
|
binning_method
|
str
|
the binning method to use.
|
'greedy'
|
points_for_mean_heterogeneity
|
int
|
number of equidistant points along the feature axis used for computing the mean heterogeneity |
30
|
Source code in effector/regional_effect_ale.py
153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 | |
plot(feature, node_idx, heterogeneity=True, centering=True, scale_x_list=None, scale_y=None, y_limits=None, dy_limits=None)
Source code in effector/regional_effect_ale.py
214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 | |
effector.regional_effect_pdp.RegionalPDP(data, model, nof_instances=10000, axis_limits=None, feature_types=None, cat_limit=10, feature_names=None, target_name=None)
Bases: RegionalPDPBase
Initialize the Regional Effect method.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
ndarray
|
the design matrix, |
required |
model
|
callable
|
the black-box model,
|
required |
axis_limits
|
Union[None, ndarray]
|
Feature effect limits along each axis
When possible, specify the axis limits manually
Their shape is |
None
|
nof_instances
|
Union[int, str]
|
Max instances to use
|
10000
|
feature_types
|
Union[list, None]
|
The feature types.
|
None
|
cat_limit
|
Union[int, None]
|
The minimum number of unique values for a feature to be considered categorical
|
10
|
feature_names
|
Union[list, None]
|
The names of the features
|
None
|
target_name
|
Union[str, None]
|
The name of the target variable
|
None
|
Methods:
| Name | Description |
|---|---|
fit |
Find subregions by minimizing the PDP-based heterogeneity. |
plot |
|
Source code in effector/regional_effect_pdp.py
59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 | |
fit(features='all', candidate_conditioning_features='all', space_partitioner='best', points_for_centering=30, points_for_mean_heterogeneity=30, use_vectorized=True)
Find subregions by minimizing the PDP-based heterogeneity.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
features
|
Union[int, str, list]
|
for which features to search for subregions
|
'all'
|
candidate_conditioning_features
|
Union[str, list]
|
list of features to consider as conditioning features
|
'all'
|
space_partitioner
|
Union[str, None]
|
the method to use for partitioning the space |
'best'
|
points_for_centering
|
int
|
number of equidistant points along the feature axis used for centering ICE plots |
30
|
points_for_mean_heterogeneity
|
int
|
number of equidistant points along the feature axis used for computing the mean heterogeneity |
30
|
use_vectorized
|
bool
|
whether to use vectorized operations for the PDP and ICE curves |
True
|
Source code in effector/regional_effect_pdp.py
136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 | |
plot(feature, node_idx, heterogeneity='ice', centering=False, nof_points=30, scale_x_list=None, scale_y=None, nof_ice=100, show_avg_output=False, y_limits=None, use_vectorized=True)
Source code in effector/regional_effect_pdp.py
219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 | |
effector.regional_effect_pdp.RegionalDerPDP(data, model, model_jac=None, nof_instances=10000, axis_limits=None, feature_types=None, cat_limit=10, feature_names=None, target_name=None)
Bases: RegionalPDPBase
Initialize the Regional Effect method.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
ndarray
|
the design matrix, |
required |
model
|
callable
|
the black-box model,
|
required |
model_jac
|
Optional[callable]
|
the black-box model's Jacobian,
|
None
|
axis_limits
|
Union[None, ndarray]
|
Feature effect limits along each axis
When possible, specify the axis limits manually
Their shape is |
None
|
nof_instances
|
Union[int, str]
|
Max instances to use
|
10000
|
feature_types
|
Union[list, None]
|
The feature types.
|
None
|
cat_limit
|
Union[int, None]
|
The minimum number of unique values for a feature to be considered categorical
|
10
|
feature_names
|
Union[list, None]
|
The names of the features
|
None
|
target_name
|
Union[str, None]
|
The name of the target variable
|
None
|
Methods:
| Name | Description |
|---|---|
fit |
Find subregions by minimizing the PDP-based heterogeneity. |
plot |
|
Source code in effector/regional_effect_pdp.py
239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 | |
fit(features='all', candidate_conditioning_features='all', space_partitioner='best', points_for_mean_heterogeneity=30, use_vectorized=True)
Find subregions by minimizing the PDP-based heterogeneity.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
features
|
Union[int, str, list]
|
for which features to search for subregions
|
'all'
|
candidate_conditioning_features
|
Union[str, list]
|
list of features to consider as conditioning features
|
'all'
|
space_partitioner
|
Union[str, None]
|
the method to use for partitioning the space |
'best'
|
points_for_mean_heterogeneity
|
int
|
number of equidistant points along the feature axis used for computing the mean heterogeneity |
30
|
use_vectorized
|
bool
|
whether to use vectorized operations for the PDP and ICE curves |
True
|
Source code in effector/regional_effect_pdp.py
322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 | |
plot(feature, node_idx=0, heterogeneity='ice', centering=False, nof_points=30, scale_x_list=None, scale_y=None, nof_ice=100, show_avg_output=False, dy_limits=None, use_vectorized=True)
Source code in effector/regional_effect_pdp.py
402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 | |
effector.regional_effect_shap.RegionalShapDP(data, model, axis_limits=None, nof_instances=1000, feature_types=None, cat_limit=10, feature_names=None, target_name=None, backend='shap')
Bases: RegionalEffectBase
Initialize the Regional Effect method.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
ndarray
|
the design matrix, |
required |
model
|
Callable
|
the black-box model,
|
required |
axis_limits
|
Optional[ndarray]
|
Feature effect limits along each axis
When possible, specify the axis limits manually
Their shape is |
None
|
nof_instances
|
Union[int, str]
|
Max instances to use
|
1000
|
feature_types
|
Optional[List[str]]
|
The feature types.
|
None
|
cat_limit
|
Optional[int]
|
The minimum number of unique values for a feature to be considered categorical
|
10
|
feature_names
|
Optional[List[str]]
|
The names of the features
|
None
|
target_name
|
Optional[str]
|
The name of the target variable
|
None
|
backend
|
str
|
Package to compute SHAP values
|
'shap'
|
Methods:
| Name | Description |
|---|---|
fit |
Fit the regional SHAP. |
plot |
Plot the regional SHAP. |
Source code in effector/regional_effect_shap.py
18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 | |
fit(features, candidate_conditioning_features='all', space_partitioner='best', binning_method='greedy', budget=512, shap_explainer_kwargs=None, shap_explanation_kwargs=None)
Fit the regional SHAP.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
features
|
Union[int, str, list]
|
the features to fit. - If set to "all", all the features will be fitted. |
required |
candidate_conditioning_features
|
Union[str, list]
|
list of features to consider as conditioning features for the candidate splits - If set to "all", all the features will be considered as conditioning features. |
'all'
|
space_partitioner
|
Union[str, Best]
|
the space partitioner to use - If set to "greedy", the greedy space partitioner will be used. |
'best'
|
binning_method
|
Union[str, Greedy, Fixed]
|
the binning method to use |
'greedy'
|
budget
|
int
|
Budget to use for the approximation. Defaults to 512. - Increasing the budget improves the approximation at the cost of slower computation. - Decrease the budget for faster computation at the cost of approximation error. |
512
|
shap_explainer_kwargs
|
Optional[dict]
|
the keyword arguments to be passed to the Code behind the sceneCheck the code that is running behind the scene before customizing |
None
|
shap_explanation_kwargs
|
Optional[dict]
|
the keyword arguments to be passed to the Code behind the sceneCheck the code that is running behind the scene before customizing |
None
|
Source code in effector/regional_effect_shap.py
133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 | |
plot(feature, node_idx, heterogeneity='shap_values', centering=True, nof_points=30, scale_x_list=None, scale_y=None, nof_shap_values='all', show_avg_output=False, y_limits=None, only_shap_values=False)
Plot the regional SHAP.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
feature
|
the feature to plot |
required | |
node_idx
|
the index of the node to plot |
required | |
heterogeneity
|
whether to plot the heterogeneity |
'shap_values'
|
|
centering
|
whether to center the SHAP values |
True
|
|
nof_points
|
number of points to plot |
30
|
|
scale_x_list
|
the list of scaling factors for the feature names |
None
|
|
scale_y
|
the scaling factor for the SHAP values |
None
|
|
nof_shap_values
|
number of SHAP values to plot |
'all'
|
|
show_avg_output
|
whether to show the average output |
False
|
|
y_limits
|
the limits of the y-axis |
None
|
|
only_shap_values
|
whether to plot only the SHAP values |
False
|
Source code in effector/regional_effect_shap.py
292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 | |