Global and Regional PDP
This tutorial is an introduction to global and regional PDP; it demonstrates how to use Effector to explain a black box function, utilizing two synthetic datasets—one with uncorrelated features and the other with correlated features.
import numpy as np
import effector
Simulation example
Data Generating Distribution
We will generate \(N=1000\) examples with \(D=3\) features. In the uncorrelated setting, all variables are uniformly distributed, i.e., \(x_i \sim \mathcal{U}(-1,1)\). In the correlated setting, we keep the distributional assumptions for \(x_1\) and \(x_2\) but define \(x_3\) such that it is identical to \(x_3\) by: \(x_3 = x_1\).
def generate_dataset_uncorrelated(N):
x1 = np.random.uniform(-1, 1, size=N)
x2 = np.random.uniform(-1, 1, size=N)
x3 = np.random.uniform(-1, 1, size=N)
return np.stack((x1, x2, x3), axis=-1)
def generate_dataset_correlated(N):
x1 = np.random.uniform(-1, 1, size=N)
x2 = np.random.uniform(-1, 1, size=N)
x3 = x1
return np.stack((x1, x2, x3), axis=-1)
# generate the dataset for the uncorrelated and correlated setting
N = 10_000
X_uncor = generate_dataset_uncorrelated(N)
X_cor = generate_dataset_correlated(N)
Black-box function
We will use the following linear model with a subgroup-specific interaction term: $$ y = 3x_1I_{x_3>0} - 3x_1I_{x_3\leq0} + x_3$$
The presence of interaction terms (\(3x_1I_{x_3>0}\), \(3x_1I_{x_3\leq0}\)) makes it impossible to define a solid ground truth effect. However, under some mild assumptions, we can agree tha
Ground truth effect (uncorrelated setting)
In the uncorrelated scenario, the effects are as follows:
-
For the feature \(x_1\), the global effect will be \(3x_1\) half of the time (when \(I_{x_3>0}\)) and \(-3x_1\) the other half (when \(3x_1I_{x_3\leq0}\)). This results in a zero global effect with high heterogeneity. The regional effect should be divided into two subregions: \(x_3>0\) and \(x_3 \leq 0\), leading to two regional effects with zero heterogeneity: \(3x_1\) and \(-3x_1\).
-
For feature \(x_2\), the global effect is zero, without heterogeneity.
-
For feature \(x_3\), there is a global effect of \(x_3\) without heterogeneity due to the last term. Depending on the feature effect method, the terms \(3x_1I_{x_3>0}\) and \(-3x_1I_{x_3\leq0}\) may also introduce some effect.
Ground truth effect (correlated setting)
In the correlated scenario, where \(x_3 = x_1\), the effects are as follows:
- For the feature \(x_1\), the global effect is \(3x_1I_{x_1>0} - 3x_1I_{x_1\leq 0}\) without heterogeneity. This is because when \(x_1>0\), \(x_3>0\), so only the term \(3x_1\) is active. Similarly, when \(x_1\leq 0\), \(x_3 \leq 0\), making the term \(-3x_1\) active.
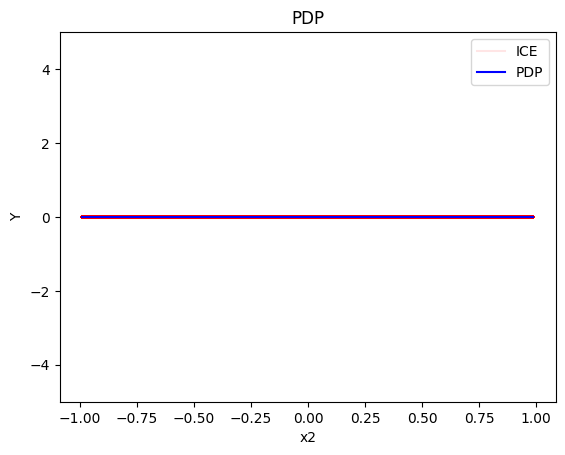
- For the feature \(x_2\), the global effect is zero, without heterogeneity.
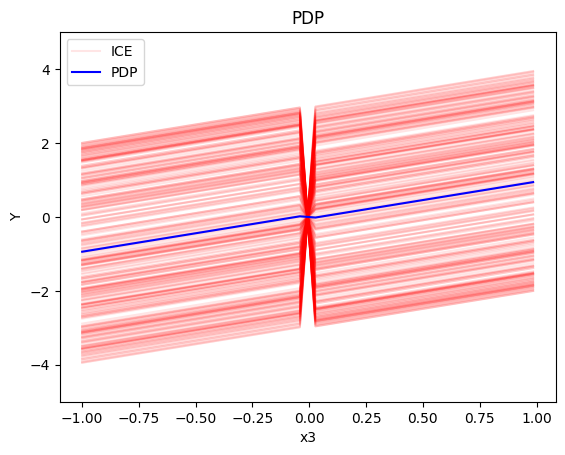
- For the feature \(x_3\), the global effect is \(x_3\).
def model(x):
f = np.where(x[:,2] > 0, 3*x[:,0] + x[:,2], -3*x[:,0] + x[:,2])
return f
def model_jac(x):
dy_dx = np.zeros_like(x)
ind1 = x[:, 2] > 0
ind2 = x[:, 2] <= 0
dy_dx[ind1, 0] = 3
dy_dx[ind2, 0] = -3
dy_dx[:, 2] = 1
return dy_dx
Y_cor = model(X_cor)
Y_uncor = model(X_uncor)
PDP definition
The PDP is defined as the average of the model's output over the entire dataset, while varying the feature of interest.:
and is approximated using the training data:
The PDP is the average of the underlying ICE curves (local effects). An ICE curve is the model's output for a single instance while varying the feature of interest. The heterogeneity is determined by how much the ICE curves differ from the PDP plot.
Uncorrelated setting
Global PDP
regional_rhale = effector.RegionalPDP(data=X_uncor, model=model, feature_names=['x1','x2','x3'], axis_limits=np.array([[-1,1],[-1,1],[-1,1]]).T)
regional_rhale.fit(heter_pcg_drop_thres=0.3, nof_candidate_splits_for_numerical=11, centering=True)
effector.binning_methods.DynamicProgramming()
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 13.68it/s]
<effector.binning_methods.DynamicProgramming at 0x7eb7497c2690>
pdp = effector.PDP(data=X_uncor, model=model, feature_names=['x1','x2','x3'], target_name="Y")
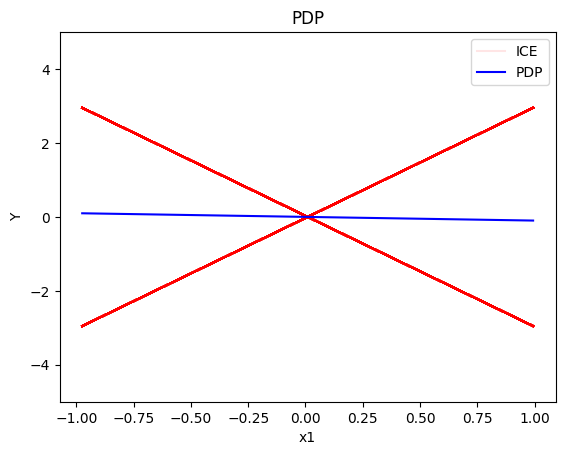
pdp.plot(feature=0, centering=True, show_avg_output=False, heterogeneity="ice", y_limits=[-5, 5])
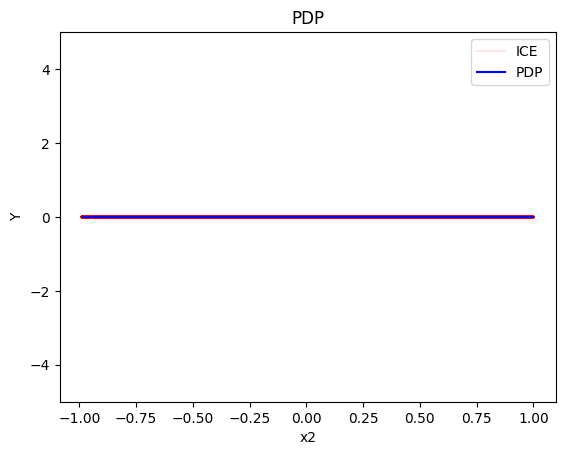
pdp.plot(feature=1, centering=True, show_avg_output=False, heterogeneity="ice", y_limits=[-5, 5])
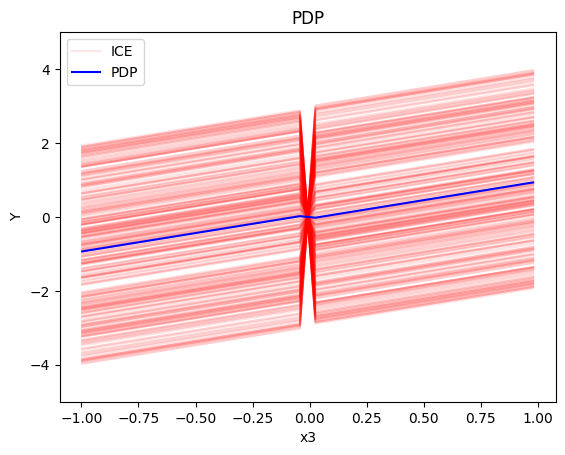
pdp.plot(feature=2, centering=True, show_avg_output=False, heterogeneity="ice", y_limits=[-5, 5])
Conclusion
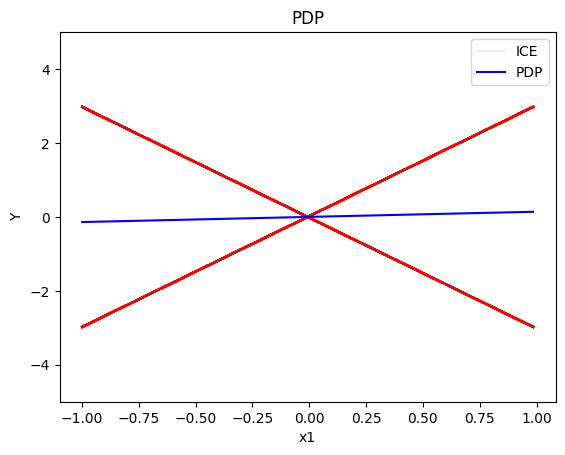
- the global effect of \(x_1\) is \(0\) with some heterogeneity due to the interaction with \(x_1\); the heterogeneity is expressed with two opposite lines; \(-3x_1\) and \(3x_1\)
- the global effect of \(x_2\) is \(0\) without heterogeneity
- the global effect of \(x_3\) is \(x_3\) with some heterogeneity due to the interaction with \(x_1\). The heterogeneity is expressed with a discontinuity around \(x_3=0\); either going from a negative offset to a positive or vice-versa.
Regional PDP
Regional PDP will search for explanations that minimize the interaction-related heterogeneity.
regional_pdp = effector.RegionalPDP(data=X_uncor, model=model, feature_names=['x1','x2','x3'], axis_limits=np.array([[-1,1],[-1,1],[-1,1]]).T)
regional_pdp.fit(features="all", heter_pcg_drop_thres=0.3, nof_candidate_splits_for_numerical=11, centering=True)
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 14.93it/s]
regional_pdp.show_partitioning(features=0)
Feature 0 - Full partition tree:
Node id: 0, name: x1, heter: 1.74 || nof_instances: 1000 || weight: 1.00
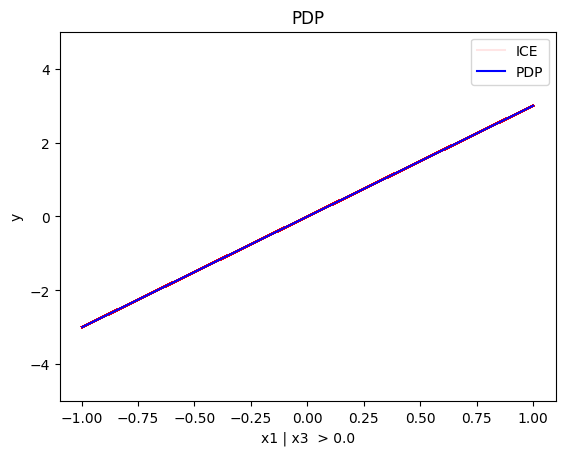
Node id: 1, name: x1 | x3 <= 0.0, heter: 0.30 || nof_instances: 515 || weight: 0.52
Node id: 2, name: x1 | x3 > 0.0, heter: 0.28 || nof_instances: 485 || weight: 0.48
--------------------------------------------------
Feature 0 - Statistics per tree level:
Level 0, heter: 1.74
Level 1, heter: 0.29 || heter drop: 1.45 (83.44%)
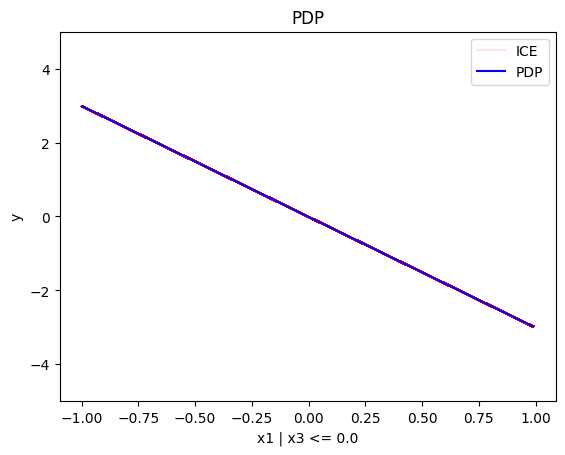
regional_pdp.plot(feature=0, node_idx=1, heterogeneity="ice", centering=True, y_limits=[-5, 5])
regional_pdp.plot(feature=0, node_idx=2, heterogeneity="ice", centering=True, y_limits=[-5, 5])
regional_pdp.show_partitioning(features=1)
Feature 1 - Full partition tree:
Node id: 0, name: x2, heter: 1.78 || nof_instances: 1000 || weight: 1.00
--------------------------------------------------
Feature 1 - Statistics per tree level:
Level 0, heter: 1.78
regional_pdp.show_partitioning(features=2)
Feature 2 - Full partition tree:
Node id: 0, name: x3, heter: 1.71 || nof_instances: 1000 || weight: 1.00
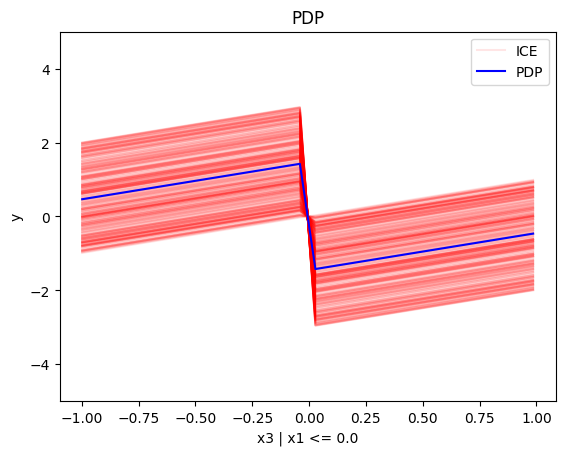
Node id: 1, name: x3 | x1 <= 0.0, heter: 0.87 || nof_instances: 506 || weight: 0.51
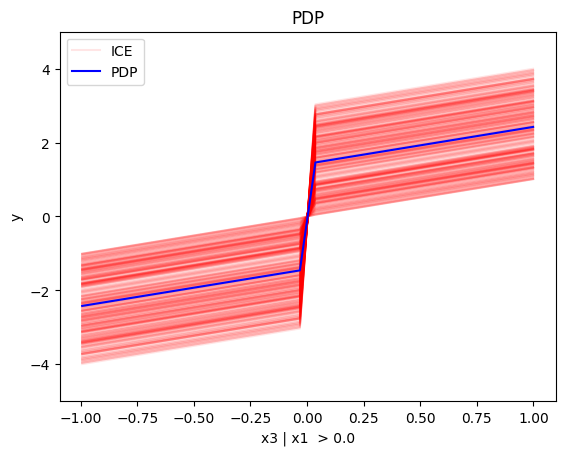
Node id: 2, name: x3 | x1 > 0.0, heter: 0.84 || nof_instances: 494 || weight: 0.49
--------------------------------------------------
Feature 2 - Statistics per tree level:
Level 0, heter: 1.71
Level 1, heter: 0.86 || heter drop: 0.85 (49.76%)
regional_pdp.plot(feature=2, node_idx=1, heterogeneity="ice", centering=True, y_limits=[-5, 5])
regional_pdp.plot(feature=2, node_idx=2, heterogeneity="ice", centering=True, y_limits=[-5, 5])
Conclusion
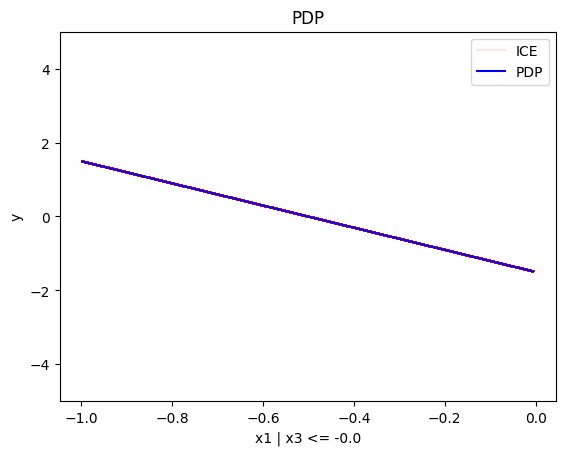
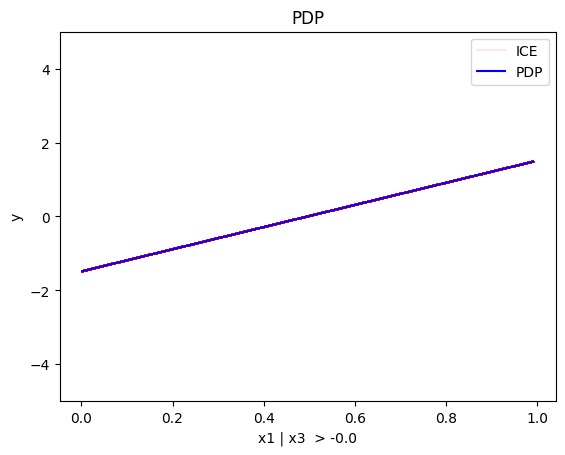
- For \(x_1\), the algorithm finds two regions, one for \(x_3 \leq 0\) and one for \(x_3 > 0\)
- when \(x_3>0\) the effect is \(3x_1\) without heterogeneity
- when \(x_3 \leq 0\), the effect is \(-3x_1\) without heterogeneity
- For \(x_2\) the algorithm does not find any subregion
- For \(x_3\), the algorithm finds two regions, there is a change in the offset:
- when \(x_1<0\) the line is \(x_3 + c^i\) in the first half and \(x_3 - c^i\) later, where \(c^i\) is \(3x_1^i\). The jump at \(x_1=0\) is therefore \(-2c^i\)
- when \(x_1>0\) the line is \(x_3 - c^i\) in the first half and \(x_3 + c^i\) later, where \(c^i\) is \(3x_1^i\). The jump at \(x_1=0\) is therefore \(2c^i\)
Correlated setting
Global PDP
pdp = effector.PDP(data=X_cor, model=model, feature_names=['x1','x2','x3'], target_name="Y")
pdp.plot(feature=0, centering=True, show_avg_output=False, heterogeneity="ice", y_limits=[-5, 5])
pdp.plot(feature=1, centering=True, show_avg_output=False, heterogeneity="ice", y_limits=[-5, 5])
pdp.plot(feature=2, centering=True, show_avg_output=False, heterogeneity="ice", y_limits=[-5, 5])
Regional-PDP
regional_pdp = effector.RegionalPDP(data=X_cor, model=model, feature_names=['x1','x2','x3'], axis_limits=np.array([[-1,1],[-1,1],[-1,1]]).T)
regional_pdp.fit(features="all", heter_pcg_drop_thres=0.4, nof_candidate_splits_for_numerical=11)
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 40.24it/s]
regional_pdp.show_partitioning(features=0)
Feature 0 - Full partition tree:
Node id: 0, name: x1, heter: 1.73 || nof_instances: 1000 || weight: 1.00
Node id: 1, name: x1 | x3 <= -0.0, heter: 0.28 || nof_instances: 483 || weight: 0.48
Node id: 2, name: x1 | x3 > -0.0, heter: 0.29 || nof_instances: 517 || weight: 0.52
--------------------------------------------------
Feature 0 - Statistics per tree level:
Level 0, heter: 1.73
Level 1, heter: 0.29 || heter drop: 1.45 (83.48%)
regional_pdp.plot(feature=0, node_idx=1, heterogeneity="ice", centering=True, y_limits=[-5, 5])
regional_pdp.plot(feature=0, node_idx=2, heterogeneity="ice", centering=True, y_limits=[-5, 5])
regional_pdp.show_partitioning(features=1)
Feature 1 - Full partition tree:
Node id: 0, name: x2, heter: 1.04 || nof_instances: 1000 || weight: 1.00
Node id: 1, name: x2 | x1 <= 0.54, heter: 0.59 || nof_instances: 759 || weight: 0.76
Node id: 2, name: x2 | x1 > 0.54, heter: 0.55 || nof_instances: 241 || weight: 0.24
--------------------------------------------------
Feature 1 - Statistics per tree level:
Level 0, heter: 1.04
Level 1, heter: 0.58 || heter drop: 0.47 (44.85%)
regional_pdp.show_partitioning(features=2)
Feature 2 - Full partition tree:
Node id: 0, name: x3, heter: 1.73 || nof_instances: 1000 || weight: 1.00
Node id: 1, name: x3 | x1 <= -0.0, heter: 0.85 || nof_instances: 483 || weight: 0.48
Node id: 2, name: x3 | x1 > -0.0, heter: 0.87 || nof_instances: 517 || weight: 0.52
--------------------------------------------------
Feature 2 - Statistics per tree level:
Level 0, heter: 1.73
Level 1, heter: 0.86 || heter drop: 0.87 (50.24%)
regional_pdp.plot(feature=2, node_idx=1, heterogeneity="ice", centering=True, y_limits=[-5, 5])
regional_pdp.plot(feature=2, node_idx=2, heterogeneity="ice", centering=True, y_limits=[-5, 5])
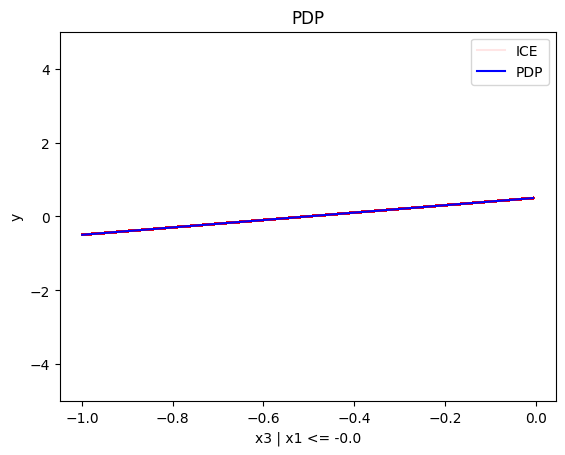
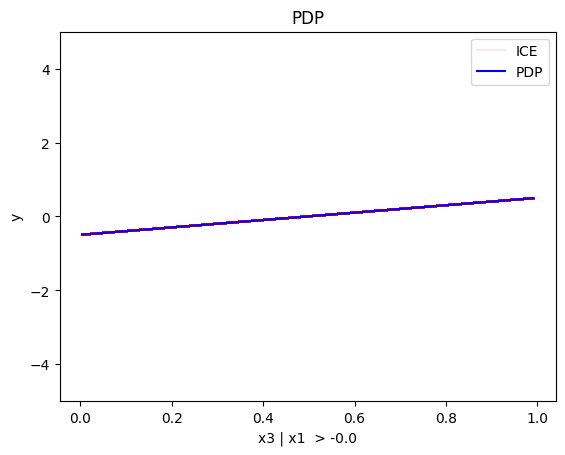
Conclusion
PDP assumes feature independence, therefore, it is not a good explanation method for the correlated case. Due to this fact, the global and regional effects are identical to the uncorrelated case.